

number of bases trimmed by bwa, that belong to non secondary and non supplementary reads. Only alignment matches(M), inserts(I), sequence matches(=) and sequence mismatches(X) are counted. number of mapped bases filtered by the CIGAR string corresponding to the read they belong to. number of processed bases that belong to

number of processed bases from reads that are neither secondary nor supplementary (flags 0x100 (256) and 0x800 (2048) are not set).

number of supplementary reads (flag 0x800 (2048) set). number of secondary reads (flag 0x100 (256) set). number of reads that failed the quality checks (flag 0x200 (512) is set). number of mapped reads with mapping quality 0. number of duplicate reads (flag 0x400 (1024) is set). number of paired reads, mapped or unmapped, that are neither secondary nor supplementary (flag 0x1 is set and flags 0x100 (256) and 0x800 (2048) are not set). number of mapped paired reads with flag 0x2 set. number of unmapped reads (flag 0x4 is set). number of mapped paired reads (flag 0x1 is set and flags 0x4 and 0x8 are not set). number of reads, paired or single, that are mapped (flag 0x4 or 0x8 not set). flag indicating whether the file is coordinate sorted (1) or not (0).įragment reads (flags 0x01 not set or flags 0x01įragment reads (flags 0x01 and 0x80 set, 0x40 not set). number of discarded reads when using -f or -F option. total number of reads in a file, excluding supplementary and secondary reads. The SN section contains a series of counts, percentages, and averages, in a similar style to The checksums are computed per alignment recordĪnd summed, meaning the checksum does not change if the input file has The CHK row contains distinct CRC32 checksums of read names, sequencesĪnd quality values. Information on the meaning of the flags is given in the SAM specification Neither is set are not counted in either category.
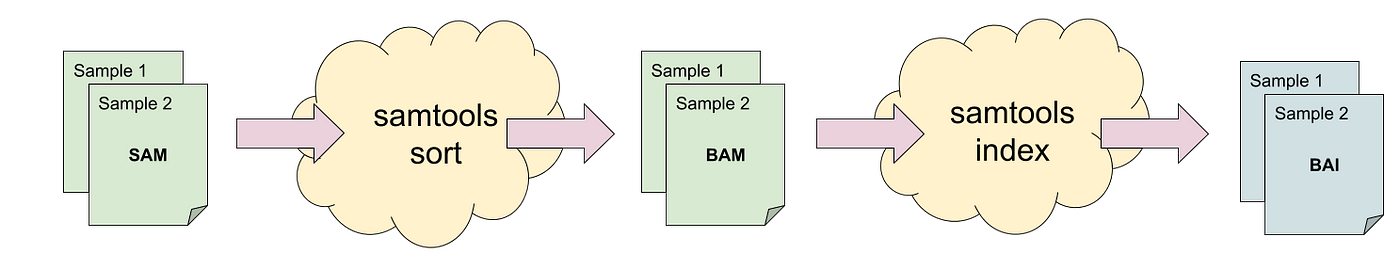
PAIRED is not set) are all “first” fragments.įor these records, the READ1 and READ2 flags are ignored. Records are put into these categories using the PAIRED (0x1), READ1 (0x40) Some of the statistics are collected for “first” or “last” Not all sections will be reported as some depend on the data beingĬoordinate sorted while others are only present when specific barcode The output can be visualized graphically using plot-bamstats.Ī summary of output sections is listed below, followed by moreĪCGT content per cycle for first fragments onlyĪCGT content per cycle for last fragments only Samtools stats collects statistics from BAM files and outputs in a text format. Samtools stats – produces comprehensive statistics from alignment file


 0 kommentar(er)
0 kommentar(er)
